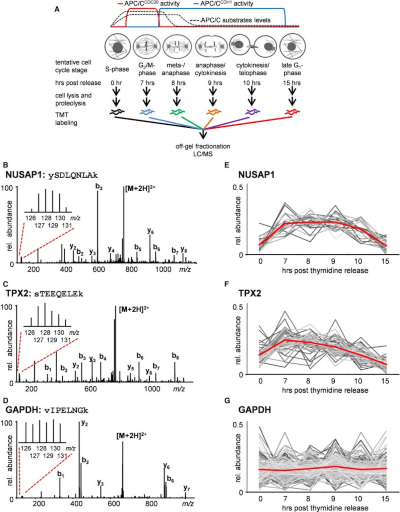
Co-regulation proteomics reveals substrates and mechanisms of APC/C-dependent degradation
Using multiplexed quantitative proteomics, we analyzed cell cycle-dependent changes of the human proteome. We identified >4,400 proteins, each with a six-point abundance profile across the cell cycle. Hypothesizing that proteins with similar abundance profiles are co-regulated, we clustered the proteins with abundance profiles most similar to known Anaphase-Promoting Complex/Cyclosome (APC/C) substrates to identify additional putative APC/C substrates. This protein profile similarity screening (PPSS) analysis resulted in a shortlist enriched in kinases and kinesins. Biochemical studies on the kinesins confirmed KIFC1, KIF18A, KIF2C, and KIF4A as APC/C substrates. Furthermore, we showed that the APC/C(CDH1)-dependent degradation of KIFC1 regulates the bipolar spindle formation and proper cell division. A targeted quantitative proteomics experiment showed that KIFC1 degradation is modulated by a stabilizing CDK1-dependent phosphorylation site within the degradation motif of KIFC1. The regulation of KIFC1 (de-)phosphorylation and degradation provides insights into the fidelity and proper ordering of substrate degradation by the APC/C during mitosis.
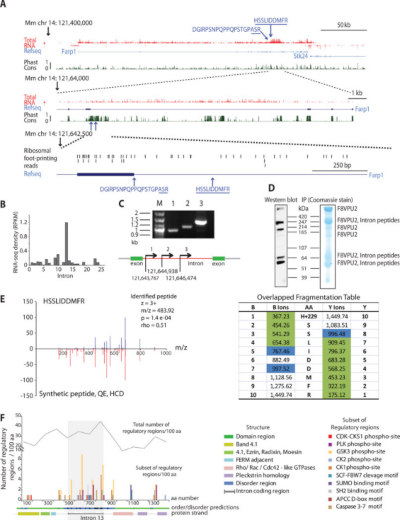
Quantitative profiling of peptides from RNAs classified as noncoding
Only a small fraction of the mammalian genome codes for messenger RNAs destined to be translated into proteins, and it is generally assumed that a large portion of transcribed sequences--including introns and several classes of noncoding RNAs (ncRNAs)--do not give rise to peptide products. A systematic examination of translation and physiological regulation of ncRNAs has not been conducted. Here we use computational methods to identify the products of non-canonical translation in mouse neurons by analysing unannotated transcripts in combination with proteomic data. This study supports the existence of non-canonical translation products from both intragenic and extragenic genomic regions, including peptides derived from antisense transcripts and introns. Moreover, the studied novel translation products exhibit temporal regulation similar to that of proteins known to be involved in neuronal activity processes. These observations highlight a potentially large and complex set of biologically regulated translational events from transcripts formerly thought to lack coding potential.
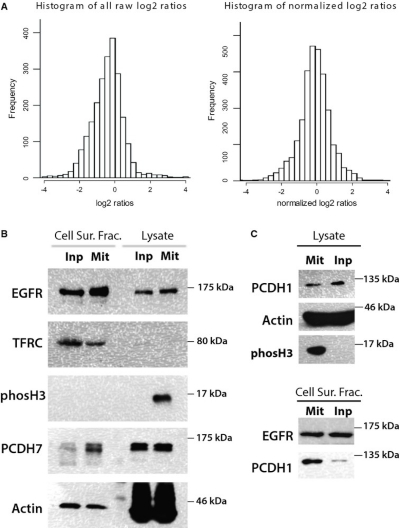
Quantitative comparison of a human cancer cell surface proteome between interphase and mitosis
The cell surface is the cellular compartment responsible for communication with the environment. The interior of mammalian cells undergoes dramatic reorganization when cells enter mitosis. These changes are triggered by activation of the CDK1 kinase and have been studied extensively. In contrast, very little is known of the cell surface changes during cell division. We undertook a quantitative proteomic comparison of cell surface-exposed proteins in human cancer cells that were tightly synchronized in mitosis or interphase. Six hundred and twenty-eight surface and surface-associated proteins in HeLa cells were identified; of these, 27 were significantly enriched at the cell surface in mitosis and 37 in interphase. Using imaging techniques, we confirmed the mitosis-selective cell surface localization of protocadherin PCDH7, a member of a family with anti-adhesive roles in embryos. We show that PCDH7 is required for development of full mitotic rounding pressure at the onset of mitosis. Our analysis provided basic information on how cell cycle progression affects the cell surface. It also provides potential pharmacodynamic biomarkers for anti-mitotic cancer chemotherapy.
Phone
Judith Steen - 617-919-2450Hanno Steen - 617-919-2629
